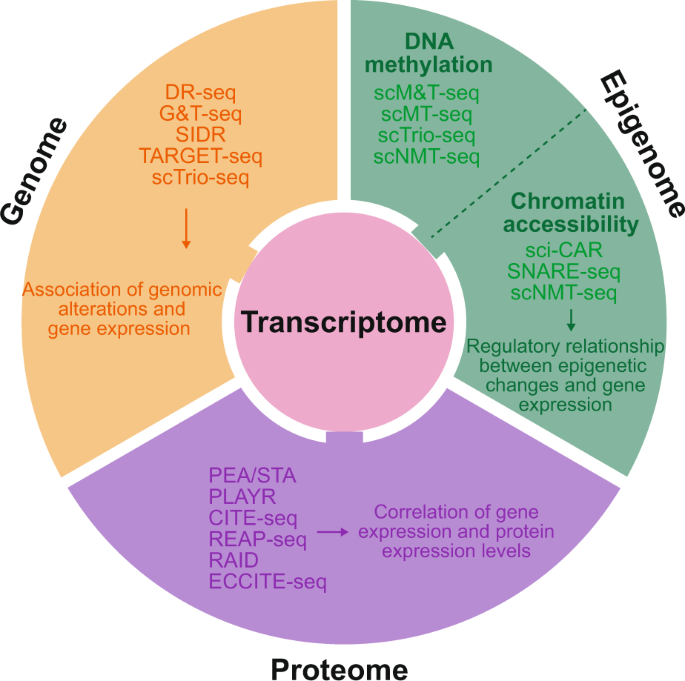
Single-cell multi-omics sequencing provides the best kind of opportunities that helps to systematically explore the diversity of cells in a definite manner. Moreover, it also has a heterogeneity that enables itself to be more comprehensive in the delineation of single cells than any other than single omics data which might be based on some kind of multichannel molecular forms.
But what is RNA Seq?
RNA-seq is a kind of single-cell sequencing technique that helps in examining the sequences of RNA as well as its quantity. This helps in using the samples of RNA in next-generation sequencing. It also helps in analyzing the various transcriptome of different gene expressions and patterns that can be easily encoded within the RNA.
But let’s know what else it is useful for and what are its applications.
What are the applications of RNA-seq?
We have already known about the single-cell RNA sequencing that proves beneficial in the treatment of various diseases. RNA-seq helps us to identify and discover the total content of cellular structures and transcriptome that includes mRNA, rRNA as well as tRNA .RNA seq helps in understand transcriptomes and connect to the various information regarding the genome with many functional expressions of protein. RNA-seq tells us about which kind of genes are turned inside the cell or what would be their level of expression and what could be the probable times they might get shut off or get activated. This provides the researchers to get a deep understanding of the biology of cells. It also helps to assess the changes that may be indicated sometimes. The most popular techniques against this thing is that using the RNA seq in transcriptionsl profiling, identification of SNP, RNA editing as well as differential analysis of gene can be beneficial.
But how does Single-cell multiomics sequencing reveal the functional regulatory landscape of early embryos?
Recent studies that have been using the technique of single-cell epigenome sequencing have been poured with huge global views of the ever-changing dynamics regarding different epigenetic layers during its period. It has been witnessed that Extensive epigenetic reprogramming usually happens during preimplantation of the embryo and its development. But it remains a bit unclear about how can the harsh epigenetic reprogramming could actually contribute to transcriptional regulation and its networking during the entire period. Therefore the technique of single-cell multi-omics sequencing (scNOMeRe-seq) was developed such that it helps in enabling the profiling of various genomes and their wide chromatin with a good amount of accessibility, DNA methylation as well as RNA expression in the same single cell. This method has also been applied in depicting the single-cell multi-omics with the map of a mouse, its preimplantation, and development. We can also discover the wide Genome DNA methylation and its remodeling that facilitates the vigorous reconstruction of many genetic lineages in the early part of embryos. Also, it has been witnessed that we can easily construct a zygotic kind of genome activation (ZGA) that is associated and linked to a network revealing the huge coordination within multiple and epigenetic layers. It also includes the various transcription factors and the repeated elements that usually prove helpful in instructing the proper zygotic kind of genome activation (ZGA). The fates of cells are linked with cis-regulatory particles that can be activated in a step-to-step manner during the post-zygotic genome activation stages. Moreover, the Trophectoderm (TE) is a kind of specific transcription factor that plays a vast role in a dual manner. It serves as beneficial in promoting the TE program while it helps in repressing and compressing the inner cell mass (ICM) program during the ICM separation. It has also been highlighted that the global DNA methylation remodeling technique actually facilitates the reconstruction of genetics in part of early embryos are driven by an asymmetric kind of cleavage. Moreover, the Allele-specific kind of DNA methylation pattern is helpful in maintaining a proper preimplantation of development that is accompanied by allele-specific associations. These associations are present between DNA methylation and definite expressions of the gene in the body that has been inherited from sperms as well as oocytes. Therefore through the integrated analyses of the dynamics in a collective manner in between the protein expression of gene and chromatin, the ZGA network is constructed with greater accessibility. The ICM is also taken altogether not only proves useful in depicting the first single-cell triple-omics during preimplantation. Therefore the technique of Single-cell multi-omics sequencing reveals the functional regulatory landscape of early embryos.